All about Polymerase Chain Reaction (PCR)
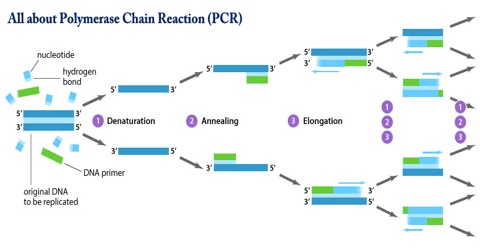
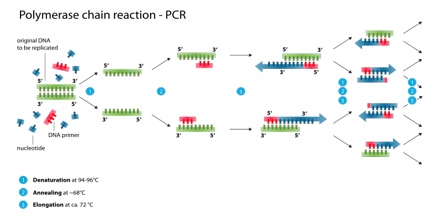
Polymerase Chain Reaction, or PCR, is a technique to make many copies of a specific DNA region in vitro or in a test tube rather than an organism. PCR is based on using the ability of DNA polymerase to synthesize new strand of DNA complementary to the offered template strand. Because DNA polymerase can add a nucleotide only onto a preexisting 3′-OH group, it needs a primer to which it can add the first nucleotide. This requirement makes it possible to delineate a specific region of template sequence that the researcher wants to amplify. At the end of the PCR reaction, the specific sequence will be accumulated in billions of copies (amplicons).
The Polymerase Chain Reaction (PCR) was originally developed in 1983 by the American biochemist Kary Mullis. He was awarded the Nobel Prize in Chemistry in 1993 for his pioneering work.
The vast majority of PCR methods rely on thermal cycling, which involves exposing the reactants to cycles of repeated heating and cooling, permitting different temperature-dependent reactions—specifically, DNA melting and enzyme-driven DNA replication—to quickly proceed many times in sequence. Primers (short DNA fragments) containing sequences complementary to the target region, along with a DNA polymerase, after which the method is named, enable selective and repeated amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the original DNA template is exponentially amplified. The simplicity of the basic principle underlying PCR means it can be extensively modified to perform a wide array of genetic manipulations. PCR is not generally considered to be a recombinant DNA method, as it does not involve cutting and pasting DNA, only amplification of existing sequences.
Typically, the goal of PCR is to make enough of the target DNA region that it can be analyzed or used in some other way. For instance, DNA amplified by PCR may be sent for sequencing, visualized by gel electrophoresis, or cloned into a plasmid for further experiments.
How does Polymerase Chain Reaction (PCR) work?
To amplify a segment of DNA using PCR, the sample is first heated so the DNA denatures, or separates into two pieces of single-stranded DNA. Next, an enzyme called “Taq polymerase” synthesizes – builds – two new strands of DNA, using the original strands as templates. This process results in the duplication of the original DNA, with each of the new molecules containing one old and one new strand of DNA. Then each of these strands can be used to create two new copies, and so on, and so on. The cycle of denaturing and synthesizing new DNA is repeated as many as 30 or 40 times, leading to more than one billion exact copies of the original DNA segment.
The entire cycling process of PCR is automated and can be completed in just a few hours. It is directed by a machine called a thermocycler, which is programmed to alter the temperature of the reaction every few minutes to allow DNA denaturing and synthesis.
Polymerase Chain Reaction (PCR) Optimization
Polymerase Chain Reaction or PCR can fail for various reasons, in part due to its sensitivity to contamination causing amplification of spurious DNA products. Because of this, a number of techniques and procedures have been developed for optimizing PCR conditions. Contamination with extraneous DNA is addressed with lab protocols and procedures that separate pre-PCR mixtures from potential DNA contaminants. This usually involves spatial separation of PCR-setup areas from areas for analysis or purification of PCR products, use of disposable plasticware, and thoroughly cleaning the work surface between reaction setups. Primer-design techniques are important in improving PCR product yield and in avoiding the formation of spurious products, and the usage of alternate buffer components or polymerase enzymes can help with amplification of long or otherwise problematic regions of DNA. Addition of reagents, such as formamide, in buffer systems may increase the specificity and yield of PCR. Computer simulations of theoretical PCR results (Electronic PCR) may be performed to assist in primer design.
The Steps of Polymerase Chain Reaction (PCR)
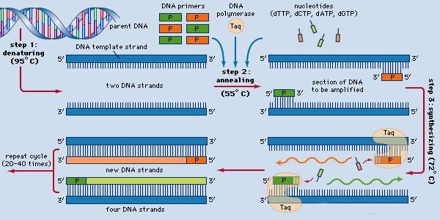
The key ingredients of a PCR reaction are Taq polymerase, primers, template DNA, and nucleotides (DNA building blocks). The ingredients are assembled in a tube, along with cofactors needed by the enzyme, and are put through repeated cycles of heating and cooling that allow DNA to be synthesized.
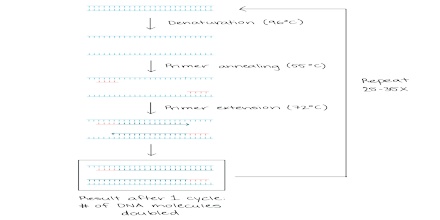
The basic steps are:
Denaturation (96°C): Heat the reaction strongly to separate, or denature, the DNA strands. This provides single-stranded template for the next step.
Annealing (555555 – 656565°C): Cool the reaction so the primers can bind to their complementary sequences on the single-stranded template DNA.
Extension (72°C): Raise the reaction temperatures so Taq polymerase extends the primers, synthesizing new strands of DNA.
Applications of Polymerase Chain Reaction (PCR)
Using PCR, a DNA sequence can be amplified millions or billions of times, producing enough DNA copies to be analyzed using other techniques. For instance, the DNA may be visualized by gel electrophoresis, sent for sequencing, or digested with restriction enzymes and cloned into a plasmid.

PCR is used in many research labs, and it also has practical applications in forensics, genetic testing, and diagnostics. For instance, PCR is used to amplify genes associated with genetic disorders from the DNA of patients (or from fetal DNA, in the case of prenatal testing). PCR can also be used to test for a bacterium or DNA virus in a patient’s body: if the pathogen is present, it may be possible to amplify regions of its DNA from a blood or tissue sample.