Plant shoots take center stage, with their fruit, blooms, and apparent structure. However, plant physiologist and computer scientist Alexander Bucksch, associate professor of Plant Biology at the University of Georgia, is interested in the component of the plant that is below the earth, the branching, reaching arms of roots and hairs that suck up water and nutrients.
The root system’s health and growth have far-reaching consequences for our future. Our capacity to grow enough food to feed the population despite a changing climate, as well as human ability to fix carbon from the atmosphere in the soil, is crucial to our existence, as well as that of other species. Bucksch believes that the characteristics of roots are the key to finding answers.
“When there is a problem in the world, humans can move. But what does the plant do?” he asked. “It says, ‘Let’s alter our genome to survive.’ It evolves.”
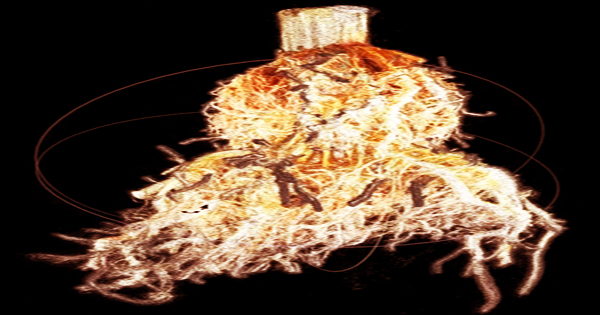
Farmers and plant breeders didn’t have a reliable means to obtain information about a plant’s root system or make judgments about the best seeds to produce deep roots until recently.
Bucksch and colleagues describe DIRT/3D (Digital Imaging of Root Characteristics), an image-based 3D root phenotyping platform that can quantify 18 architectural traits from mature field-grown maize root crowns dug using the Shovelomics approach, in a work published this month in Plant Physiology.
In their tests, the system accurately calculated all characteristics for 12 different maize genotypes, including the distance between whorls and the number, angles, and diameters of nodal roots, with an 84 percent agreement with manual measurements. The research is funded by the Advanced Research Projects Agency-Energy (ARPA-E) ROOTS program and a National Science Foundation CAREER award (NSF).
“This technology will make it easier to analyze and understand what roots are doing in real field environments, and therefore will make it easier to breed future crops to meet human needs,” said Jonathan Lynch, Distinguished Professor of Plant Science and co-author, whose research focuses on understanding the basis of plant adaptation to drought and low soil fertility.
DIRT/3D has a motorized camera system that captures 2,000 pictures per root from all angles. It utilizes a cluster of ten Raspberry Pi microcomputers to synchronize picture collection from ten cameras, then transmits the data to the CyVerse Data Store, a national cyberinfrastructure for academic researchers, for 3D reconstruction.
According to Bucksch, the method creates a 3D point cloud that depicts each root node and whorl as a “digital twin of the root system” that can be analyzed, saved, and compared. Data gathering takes only a few minutes, similar to that of an MRI or X-Ray equipment.
However, instead of costing half a million dollars to create, the rig just costs a few thousand dollars, making the technology scalable enough to do high-throughput tests on thousands of specimens, which is required to develop new agricultural plants for farmers. The 3D scanner, on the other hand, is allowing fundamental science and addressing the problem of pre-selection bias in plant biology due to sample constraints.
“Biologists primarily look at the one root structure that is most commonly what we call the dominant root phenotype,” Bucksch explained. “But people forgot about all of the other phenotypes. They might have a function and a role to fulfill. But we just call it noise,” Bucksch said. “Our system will look into that noise in 3D and see what functions these roots might have.”
Individuals who use DIRT/3D to photograph roots will soon be able to upload their data to PlantIT, a website that can do the same analysis Bucksch and his collaborators describe in their new work, giving data on a wide variety of characteristics from juvenile nodal root length to root system eccentricity. Researchers and breeders can use this information to compare the root systems of plants grown from the same or different seeds.
Massive number-crunching skills behind the scenes enable the framework. The Texas Advanced Computing Center (TACC), which gets enormous volumes of data from the CyVerse Cyberinfrastructure for computing, provides several services.
Though imaging a root crown takes only five minutes, the data processing required to build the point cloud and quantify the characteristics takes many hours and requires multiple processors to work in parallel. Bucksch utilizes an allocation from the Extreme Science and Engineering Discovery Environment (XSEDE) to facilitate his research and run the public DIRT/2D and DIRT/3D servers on TACC’s NSF-funded Stampede2 supercomputer.
DIRT/3D is a development of a previous 2D version of the program that could extract root information with only a cell phone camera. DIRT/2D has shown to be a valuable tool for the field since its introduction in 2016. It is used by hundreds of plant scientists throughout the world, including researchers at major agribusinesses.
The research is part of ARPA-ROOTS E’s program, which aims to develop innovative technologies that help plants store more carbon in their soil and root systems.
“The DIRT/3D platform enables researchers to identify novel root traits in crops, and breed plants with deeper, more extensive roots,” said ARPA-E ROOTS Program Director Dr. David Babson. “Climate change mitigation and resilience will be aided by the development of such technology, which will also provide farmers with tools to reduce costs and enhance agricultural yield. We’re looking forward to seeing how far the team from PSU and UGA has progressed throughout the course of their award.”
The technique has resulted in the identification of many genes involved in root characteristics. Bucksch points to a recent study on Striga hermanthica resistance in sorghum as an example of the type of result he wants for DIRT/3D users. Striga, a parasitic plant, decimates sorghum yields across vast swaths of Africa.
Dorota Kawa, a postdoctoral researcher at UC Davis, discovered that some strains of sorghum have Striga-resistant roots. She used DIRT/2D to extract characteristics from these roots, which she then linked to genes that control the release of chemicals in the roots that cause Striga germination in plants.
DIRT3D enhances the quality of DIRT/2D root characterizations by capturing characteristics that are only visible when scanned in three dimensions.
Droughts, rising temperatures, low soil fertility, and the need to grow food in fewer greenhouse-gas-producing methods are all likely to exacerbate farmers’ problems in the future years. Roots that have been modified to these future circumstances will assist to alleviate food shortages.
“The potential, with DIRT/3D, is helping us live on a hotter planet and managing to have enough food,” Bucksch said. “That is always the elephant in the room. There could be a point where this planet can’t produce enough food for everybody anymore, and I hope we, as a science community, can avoid this point by developing better drought-adapted and CO2 sequestering plants.”