Many strategies have been developed to trap and monitor single proteins over time in order to discover the molecular mechanisms of these critical nanomachines. Single-protein sensing with nanopores is especially appealing because it enables label-free high-bandwidth detection based on ion currents.
Researchers from the technical universities of Delft and Munich have developed a new type of molecular trap that can hold a single protein in place for hours to study its natural behavior—a million times longer than previously possible. The new NEOtrap technique allows scientists to study the vibrant nature of proteins using electrical currents, which could spark innovation in biomedicine, biotechnology, and other fields.
Even though proteins are essential to life—for example, providing you with vision and neural connections to read this text—the process by which they change shape is still unknown. A team led by Cees Dekker at the Delft University of Technology developed a new technique called the Nanopore Electro-Osmotic trap (NEOtrap), which was published on Monday 30 August in Nature Nanotechnology, to study individual protein molecules for much longer than was previously possible. The NEOtrap enables researchers to track how single proteins change shape over time.
Researchers invented a new type of molecular trap that can hold a single protein in place for hours to study its natural behavior a million times longer than before.
Like a cork on a bottle
The NEOtrap brings together two nanotechnologies: solid-state nanopores and DNA origami. Nanopores are tiny holes that scientists use to detect single molecules like proteins. Proteins can only be recorded briefly because they normally pass through the small hole in microseconds. The researchers can lock the protein in place for hours by sealing the nanohole with a nanoscale ball made entirely of DNA (! ), similar to how a cork seals a bottle of wine. Hendrik Dietz and his colleagues at the Technical University of Munich created this nano-ball using “DNA origami,” a technique that mimics macroscale origami folding but with nanoscale DNA strands rather than paper.
Sonja Schmid, the paper’s lead author who developed the NEOtrap as a postdoc in Dekker’s lab, explains: “This DNA-origami nano-ball acts like a sponge, sucking water through the nanopore and attracting and trapping a single protein. This means we’ll be able to study that protein for a very long time. In this paper, we show that we can distinguish between different types of proteins, as well as different functional shapes of the same protein.”
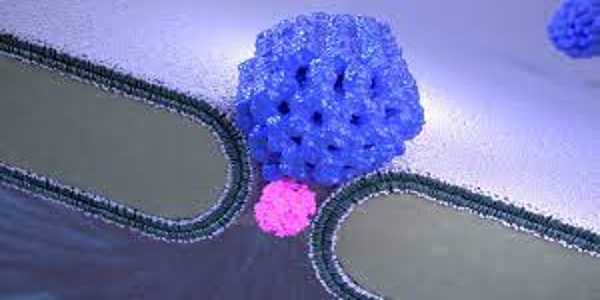
Radical advancement in the field
Cees Dekker continues: “This new technique is truly revolutionary—an anonymous reviewer of our paper called it “one of the most radical advancements in the field of nanopore sensing.” Notably, unlike previous techniques, the NEOtrap allows us to trap a single native protein without modifying the molecule of interest. This technique, for example, can assist researchers in discovering the underlying mechanism of enzymes and other important proteins that change shape to facilitate chemical reactions.”
The NEOtrap allows scientists all over the world to conduct entirely new experiments, which have the potential to reveal previously unknown functional features of proteins and thus spark innovation in biomedicine, biotechnology, and other fields. In the coming years, Schmid (who has now established her own lab at Wageningen) and Dekker intend to conduct numerous follow-up studies on the dynamics of single proteins.
Techniques for studying and manipulating single molecules of DNA and other biopolymers have been developed in recent years. A single DNA molecule is bound at one end to a glass surface and at the other to a magnetic microbead in one such technique, the magnetic trap. Small magnets with controllable position and rotation pull on and rotate the microbead. This provides a straightforward method for stretching and twisting the molecule.
To keep water levels low in water-sensitive reagents, molecular traps are used in Oligo Synthesis reagents. The Molecular Traps can be used in Oligo Synthesis Reagents for an extended period of time without the packing material or casing degrading. The amount of moisture the molecular trap has been exposed to is the determining factor for how long to use a Molecular Trap on oligo synthesis reagents before changing to a new trap. The shorter the life of the molecular trap, the wetter the solvents.